基于转录组测序的斧翅沙芥( Pugionium dolabratum )干旱胁迫应答基因表达分析
Transcriptiomic analysis of drought response genes in Pugionium dolabratum
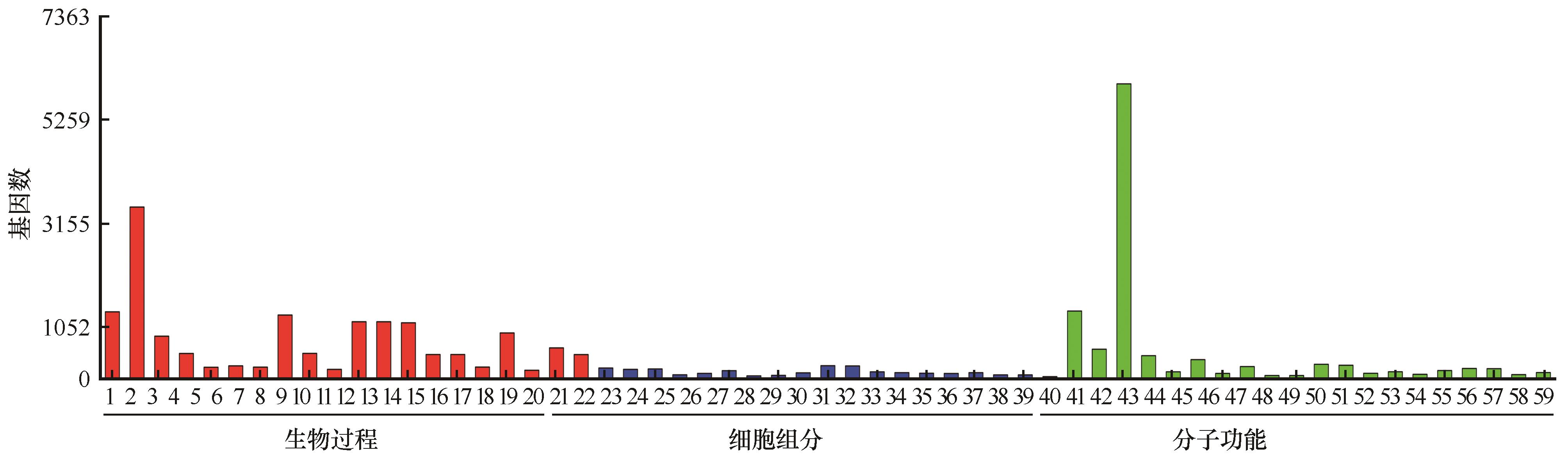
1:氧化还原过程;2:单一生物代谢过程;3:脂质代谢过程;4:单羧酸代谢过程;5:光合作用;6:单羧酸生物合成过程;7:脂肪酸代谢过程;8:单一生物合成过程;9:脂质生物合成过程;10:脂肪酸生物合成过程;11:氧乙酸代谢过程;12:有机酸代谢过程;13:羧酸代谢过程;14:有机酸生物合成过程;15:羧酸生物合成过程;16:细胞醛代谢过程;17:碳水化合物代谢过程;18:甘油醛-3-磷酸代谢过程;19:细胞脂质代谢过程;20:化学反应;21:类囊体;22:光合膜;23:类囊体部分;24:光系统II放氧复合物;25:类囊体膜;26:光系统;27:光系统I;28:质外体;29:光系统II;30:质体;31:叶绿体;32:外包裹结构;33:叶绿体部分;34:细胞壁;35:氧化还原酶复合物;36:质体部分;37:叶绿体包膜;38:质体包膜;39:系统I反应中心;40:氧化还原酶活性;41:辅因子结合;42:催化活性;43:辅酶结合;44:磷酸吡哆醛结合;45:裂解酶活性;46:单加氧酶活性;47:氧化还原酶活性,作用于成对供体,分子氧掺入或减少;48:转氨酶活性;49:转移酶活性,转移含氮基团;50:氧化还原酶活性,作用于供体的CH-OH基团;51:氧化还原酶活性,作用于供体的CH-OH基团,NAD或NADP作为受体;52:氧化还原酶活性,作用于供体的醛基;53:抗氧化活性;54:氧化还原酶活性,作用于供体的醛基,NAD或NADP作为受体;55:碳碳裂解酶活性;56:四吡咯结合;57:亚铁原卟啉结合;58:氧化还原酶活性,作用于单供体并结合分子氧;59:羧基裂解酶活性 1: oxidation-reduction process; 2: single-organism metabolic process; 3: lipid metabolic process; 4: monocarboxylic acid metabolic process; 5: photosynthesis; 6: monocarboxylic acid biosynthetic process; 7: fatty acid metabolic process; 8: single-organism biosynthetic process; 9: lipid biosynthetic process; 10: fatty acid biosynthetic process; 11: oxoacid metabolic process; 12: organic acid metabolic process; 13: carboxylic acid metabolic process; 14: organic acid biosynthetic process; 15: carboxylic acid biosynthetic process; 16: cellular aldehyde metabolic process; 17: carbohydrate metabolic process; 18: glyceraldehyde-3-phosphate metabolic process; 19: cellular lipid metabolic process; 20: response to chemical; 21: thylakoid; 22: photosynthetic membrane; 23: thylakoid part; 24: photosystem II oxygen evolving complex; 25: thylakoid membrane; 26: photosystem; 27: photosystem I; 28: apoplast; 29: photosystem II; 30: plastid; 31: chloroplast; 32: external encapsulating structure; 33: chloroplast part; 34: cell wall; 35: oxidoreductase complex; 36: plastid part; 37: chloroplast envelope; 38: plastid envelope; 39: photosystem I reaction center; 40: oxidoreductase activity; 41: cofactor binding; 42: catalytic activity; 43: coenzyme binding; 44: pyridoxal phosphate binding; 45: lyase activity; 46: monooxygenase activity; 47: oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen; 48: transaminase activity; 49: transferase activity, transferring nitrogenous groups; 50: oxidoreductase activity, acting on CH-OH group of donors; 51: oxidoreductase activity, acting on the CH-OH group of donors, NAD or NADP as acceptor; 52: oxidoreductase activity, acting on the aldehyde group of donors; 53: antioxidant activity; 54: oxidoreductase activity, acting on the aldehyde group of donors, NAD or NADP as acceptor; 55: carbon-carbon lyase activity; 56: tetrapyrrole binding; 57: heme binding; 58: oxidoreductase activity, acting on single donors with incorporation of molecular oxygen; 59: carboxy-lyase activity